✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
MinElute Gel Extraction Kit (50)
Cat. No. / ID: 28604
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Very small elution volumes
- Fast procedure and easy handling
- High, reproducible recoveries
- Gel loading dye for convenient sample analysis
Product Details
The MinElute Gel Extraction Kit provides spin columns, buffers, and collection tubes for silica-membrane-based purification of DNA fragments of 70 bp – 4 kb from up to 400 mg gel slices. The spin columns are designed to allow elution in very small volumes (as little as 10 µL), delivering high yields of highly concentrated DNA. An integrated pH indicator allows easy determination of the optimal pH for DNA binding to the spin column. DNA fragments purified with the MinElute system are ready for direct use in all applications, including sequencing, microarray analysis, ligation and transformation, restriction digestion, labeling, microinjection, PCR, and in vitro transcription. The MinElute Gel Extraction Kit can be automated on the QIAcube Connect.
For optimal results it is recommended to use this product together with QIAvac 24 Plus.
Performance
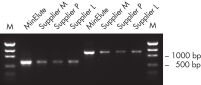
The MinElute Gel Extraction Kit removes nucleotides, enzymes, salts, agarose, ethidium bromide, and other impurities from DNA samples, delivering highly concentrated DNA suitable for a variety of downstream applications (see figure " Higher DNA concentrations").
The MinElute Gel Extraction Kit provides spin columns for gel extraction. Using a microcentrifuge or vacuum manifold, high concentration DNA of 70 bp – 4 kb is quickly purified. (DNA fragments from 4 kb – 10 kb should be purified using the QIAquick Gel Extraction Kit and DNA fragments smaller than 70 bp or larger than 10 kb should be extracted with the QIAEX II Gel Extraction System.)
See figures
Principle
MinElute Kits contain a silica membrane assembly for binding of DNA in high-salt buffer and elution with low-salt buffer or water. Silica-membrane technology eliminates the problems and inconvenience associated with loose resins and slurries. Specialized binding buffers are optimized for specific applications and promote selective adsorption of DNA molecules within particular size ranges.
Gel loading dye
To enable faster and more convenient sample processing and analysis, gel loading dye is provided. GelPilot Loading Dye contains three tracking dyes (xylene cyanol, bromophenol blue, and orange G) to facilitate the optimization of agarose gel run time and prevent smaller DNA fragments migrating too far (see figure " GelPilot Loading Dye").
See figures
Procedure
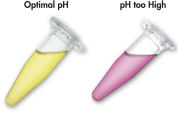
The MinElute system uses a simple bind-wash-elute procedure (see flowchart " MinElute procedure"). Gel slices are dissolved in a buffer containing a pH indicator, allowing easy determination of the optimal pH for DNA binding (see figure " pH Indicator Dye"), and the mixture is applied to the MinElute spin column. Nucleic acids adsorb to the silica-gel membrane in the high-salt conditions provided by the buffer. Impurities are washed away and pure DNA is eluted with a small volume of low-salt buffer provided or water, ready to use in subsequent applications.
Handling
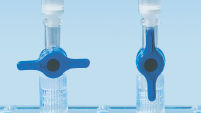
MinElute spin columns are designed to provide two convenient handling options. The spin columns fit into a conventional table-top microcentrifuge or onto any vacuum manifold with luer connectors, such as the QIAvac 24 Plus with QIAvac Luer Adapters. The MinElute Gel Extraction Kit, in addition to other QIAGEN spin-column-based kits, can be fully automated on the QIAcube Connect, enabling increased productivity and standardization of results (see figures "Spin column handling options A, B, C, D and QIAcube Connect").
See figures
Applications
DNA fragments purified with the MinElute or QIAquick System are ready for direct use in all applications, including:
- Sequencing, including next generation sequencing
- Microarray analysis
- Ligation and transformation
- Restriction digestion
- Labeling
Supporting data and figures
Spin column handling options — C.

Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 5 µg |
| Elution volume | 10 µL |
| Fragment size | 70 bp – 4 kb |
| Sample type: applications | DNA: PCR reactions |
| Technology | Gel extraction |
| Recovery: oligonucleotides dsDNA | Recovery: dsDNA fragments |
| Format | Tube |
| Processing | Manual |