Type-it Fast SNP Probe PCR Kit
For accurate and reliable SNP genotyping using TaqMan® or TaqMan® MGB probes
For accurate and reliable SNP genotyping using TaqMan® or TaqMan® MGB probes
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 206045
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
The Type-it Fast SNP Probe PCR Kit consistently ensures highly accurate SNP genotyping.
The Type-it Fast SNP Genotyping PCR Master Mix is based on highly specific HotStarTaq Plus DNA Polymerase and a newly developed SNP genotyping PCR buffer system, both of which enable highly specific probe binding and consistently strong fluorescent signals. Compared to other commercially available SNP genotyping master mix chemistries, wider and clearer separation of allele clusters is obtained (see figure " Highly specific probe binding").
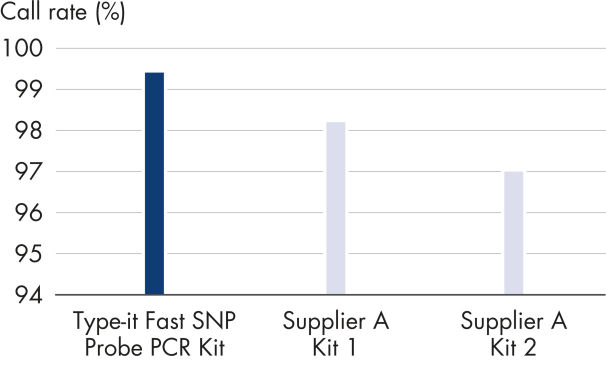
Even with challenging targets or difficult SNP loci, the Type-it Fast SNP Probe PCR Kit provides reliable SNP genotyping — well-separated allele clusters and tight clustering of identical alleles are achieved, resulting in high call rates (see figure " Successful automated allele calling"). The Type-it Fast SNP Probe PCR Kit outperformed kits tested from other suppliers, ensuring tight clustering of alleles — even with 1 ng of template DNA (see figure " Reliable SNP genotyping with small amounts of template").
The Type-it Fast SNP Probe PCR Kit is available in a convenient master mix format consisting of highly specific HotStarTaq Plus Polymerase and a unique SNP genotyping buffer system designed for fast and efficient amplification. The unique kit components provide tight allele clustering and outstanding separation to ensure high call rates, enabling reproducible and accurate results (see table).
HotStarTaq Plus DNA Polymerase, provided in the master mix, which ensures highly specific amplification, even with low template amounts. HotStarTaq Plus DNA Polymerase is provided in an inactive state with no polymerase activity at ambient temperatures. This prevents the formation of misprimed products and primer dimers at low temperatures.
The Type-it Fast SNP PCR Buffer, also included in the master mix, is specifically designed for fast-cycling SNP genotyping using sequence specific 5'-nuclease probes. The unique composition of the Type-it Fast SNP PCR Buffer ensures highly stringent and specific binding of the allele-specific probe (match probe). This is due to the altered melting behaviour of the probes resulting in a narrower probe melting temperature window. Based on the original QIAGEN PCR Buffer, this innovative formulation enables a high ratio of specific-to-nonspecific primer binding during the short annealing step of every PCR cycle. Owing to a uniquely balanced combination of KCl and (NH4)2SO4, the PCR buffer provides stringent primer annealing conditions over a wider range of annealing temperatures and Mg2+ concentrations than conventional PCR buffers. Optimization of PCR by varying the annealing temperature or the Mg2+ concentration is dramatically reduced and often not required.
Additives in the Type-it Fast SNP PCR buffer, such as Q-Solution, provide reaction conditions for amplification of difficult genomic regions and difficult SNP loci. Innovative Q-Bond technology (for fast cycling) allows SNP genotyping results to be achieved faster (see figure " Mechanism of fast cycling during annealing"). The master mix also contains ROX dye at a concentration compatible with SNP genotyping on all instruments from Applied Biosystems. However, ROX dye is also compatible with other instruments not requiring ROX as a passive reference dye (see table).
| Kit contents | Features |
|---|---|
| 2x Master Mix format* | Developed for SNP genotyping with TaqMan® MGB probes Suitable for use with all cyclers suitable for SNP genotyping |
| HotStarTaq Plus DNA Polymerase | Fast and easy reaction setup at room temperature Highly specific amplification, even with low template amounts |
| Type-it Fast SNP Probe PCR Buffer | Wide separation of genotype clusters Tight allele clusters and high allele calling rates Increased specificity in probe binding Fast cycling due to Q-Bond Molecule |
| Q-Solution | Tighter clusters and higher signals in scatter plot analysis for highly GC-rich SNP loci Can further improve suboptimal allele calling |
The Type-it Fast SNP Probe PCR Kit is functionally verified with commercially available SNP genotyping assays and is compatible with TaqMan® MGB Probes, as well as user-developed probe-based assays consisting of TaqMan® MGB, TaqMan®, or other dual-labeled probes. The kit includes streamlined, preoptimized protocols for fast and reliable analysis.
The Type-it Fast SNP Probe PCR Master Mix provides reaction conditions for fast-cycling PCR using sequence-specific probes. The buffer includes proprietary Q-Bond Molecule, which allows short cycling times on standard cyclers and on fast cyclers with rapid ramping rates. The fast-cycling procedure increases throughput by reducing PCR run times by up to 40% (see figure " Genotyping workflow").
The kit is provided in a ready-to-use, preoptimized master mix format for greater convenience. Use of a master mix saves time, simplifies handling for reaction setup, and increases reproducibility by eliminating many possible sources of pipetting errors and contamination — pipetting steps are minimized and tedious calculations are eliminated. HotStarTaq Plus DNA Polymerase (included in the master mix) is activated by a 5-minute, 95°C incubation step, which can easily be incorporated into existing thermal cycling programs. Room-temperature reaction setup using the master mix is fast and easy.
The Type-it Fast SNP Probe PCR Kit is optimized for amplification of SNP PCR assays on any suitable standard and fast ramping cycler compatible with optical PCR plates that fit into real-time PCR instruments used for fluorescence plate read analysis (see table).
| Cyclers | Model |
|---|---|
| Real-time PCR cyclers | QIAGEN: Rotor-Gene Q ABI PRISM 7900 (all series) ABI StepOne and StepOne Plus Applied Biosystems 7500 (all series) Applied Biosystems ViiA 7 Bio-Rad: iCycler iQ Bio-Rad: CFX series Roche: LightCycler 480 Roche: LightCycler Nano Agilent: Mx3000P and Mx3005P |
| Standard cyclers | All cyclers in standard-ramping modes (e.g., GeneAmp 9700) All cyclers in fast-ramping modes (e.g., GeneAmp 9800) |
The Type-it Fast SNP Probe PCR Kit is used to detect SNPs using TaqMan® MGB probes and can be used in various fields of research, including:
Average automated allele call rates for a panel of different DNAs were analyzed using 6 different TaqMan® MGB-based SNP genotyping assays and 1 ng of genomic DNA from each of the 60 samples. PCR was performed with the indicated products in 384-well plates in a 5 μl reaction volume. Allelic discrimination plate read was performed on an Applied Biosystems 7900HT instrument. The Type-it Fast SNP Probe PCR Kit consistently resulted in the highest call rates and the lowest error rates.
| Features | Specifications |
|---|---|
| Applications | Probe-based SNP Genotyping |
| Product use | Functionally validated and developed for reliable SNP Genotyping |
| Mastermix | Yes |
| Real-time or endpoint | Both |
| With or without ROX | ROX included in Master Mix |
| With/without hotstart | With |
| Sample/target type | Genomic DNA |
| Enzyme activity | 5'-> 3' Exonuclease activity |
| Sequence specific Probe | TaqMan® Genotyping Assays, TaqMan® or TaqMan® MGB probes |
| Reaction type | PCR amplification |