✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
dPCR PanCancer Kit BRAF V600 FAM (200)
Cat. No. / ID: 250284
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Parallel detection of hallmark mutations with frequencies as low as 0.1%
- Reference assay quantifies genome copies and check for dPCR efficiency
- Reference gene and hallmark mutations detected in a single run using two different dyes
- Simple and fast dPCR protocol on the QIAcuity
- Compatible with QIAGEN sample preparation kits and instruments
Product Details
The dPCR PanCancer Kits have been designed for various cancer-relevant target genes. Using a single assay, the kits can detect multiple mutations or deletions in a target gene. The dPCR PanCancer Kit BRAF V600 FAM can detect eight key mutations in BRAF V600, and the dPCR PanCancer Kit EGFRex19del FAM can detect 23 deletions in exon 19 of the EGFR gene.
Each dPCR PanCancer Kit consists of two boxes: one box with the specific PanCancer Assay with FAM-labeled gene-specific primers and a HEX-labeled reference gene to check for PCR efficiency, and the other with the QIAcuity Master Mix (see figure " dPCR PanCancer Kit concept"). The primers and probes come pre-mixed in the right concentrations.
The assay is optimized for QIAcuity Nanoplate 26k 24-well, but other nanoplate formats are also possible. The easy-to-use dPCR workflow on the QIAcuity allows fast, precise and cost-efficient screening of your samples.
See figures
Performance
High sensitivity in mutation detection
The dPCR PanCancer Kits have been developed to meet the need for precise and time-efficient screening of cancer samples in your research projects. The assays can detect multiple mutations in parallel, down to 0.1% mutation frequency, in various sample types used in cancer research.
Parallel detection of multiple hallmark mutations and reference gene
The QIAcuity Nanoplate 26k 24-well allows the analysis of 24 samples in parallel. Each assay detects several cancer-relevant mutations in parallel within a specific target gene in a single channel. As the reference assay for AP3B1 comes with a different dye than the assay for the mutations, the readout can be done using two different channels on the QIAcuity but in the same reaction well, saving precious samples and allowing more samples to be run on one plate (see figures " Accurate detection of BRAF V600E mutations in DNA reference standard using the dPCR PanCancer Kit BRAF V600 FAM" and " Accurate detection of EGFR exon19 deletion in DNA reference standard using the dPCR PanCancer Kit EGFRex19del FAM).
Compatible with multiple sample types and sample preparation methods
The dPCR PanCancer Kits are compatible with QIAGEN sample collection devices, preparation kits and instruments, allowing you to meet diverse throughput, reproducibility and sensitivity needs. Whether you isolate your DNA manually using a QIAGEN kit, such as the QIAamp Circulating Nucleic Acid Kit for cfDNA, QIAamp DNA Blood Kits for blood DNA or QIAamp DNA FFPE Advanced Kit for FFPE DNA, or perform automated DNA isolation using the EZ2 Connect or QIAsymphony SP, the dPCR PanCancer Kits work with all of them.
See figures
Principle
The dPCR PanCancer Kits – BRAF V600 FAM and EGFRex19del FAM – detect multiple hallmark mutations in the serine/threonine protein kinase B-Raf encoding gene BRAF and the epidermal growth factor receptor encoding gene EGFR, respectively.
The assays are designed for use with the QIAcuity Master Mix and the QIAcuity Digital PCR System. Each assay involves endpoint PCR amplification targeting specific genetic regions of the relevant cancer gene. The amplified product is detected using target-specific fluorescent hydrolysis probes, enhancing the assay's specificity.
Below is a detailed overview of the covered mutations with the unique COSMIC database v99 identifiers. Additionally, each dPCR PanCancer Kit incorporates a reference assay that quantifies the human single-copy gene AP3B1, providing a benchmark for determining genome copies in the sample. While the recommended setup involves a QIAcuity Nanoplate 26k 24-well, these assays are also compatible with other QIAcuity Nanoplate types, offering versatility in experimental configurations. You can also download a table with a summary of positive control templates for the BRAF and EGFR PanCancer Assays (see Resources section).
Learn the principle behind QIAcuity nanoplate-based digital PCR here.
Procedure
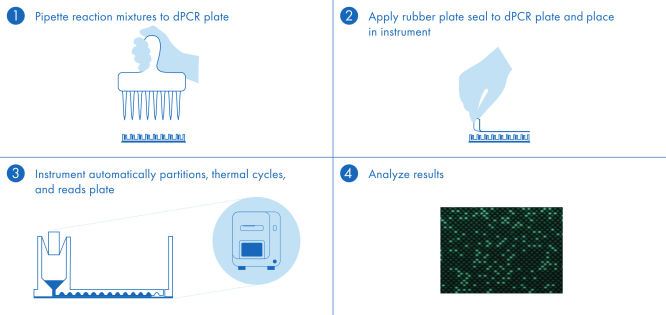
The dPCR PanCancer Kit protocol is simple and can be performed in any laboratory with the QIAcuity dPCR instrument. Following the instructions in the user manual and the Quick Start Protocol, the user prepares the reaction mix and pipettes the mix into the nanoplate. We recommend using the 26k nanoplate and keeping the software up to date. Isolated DNA is added to the ready-to-use QIAcuity Probe Master Mix and nuclease-free water. This mix is aliquoted into each well of the dPCR pre-plate, which contains pre-dispensed sets of primers and hydrolysis probes. The reaction mixtures are transferred from the pre-plate to the dPCR nanoplate wells, which are then sealed and transferred to the QIAcuity dPCR instrument (see figure " A simple and rapid plate-based workflow"). The partitioning, cycling and imaging steps are fully automated by the QIAcuity dPCR instrument following the preset parameters. Depending on the cycling protocol, the results of the dPCR run can be analyzed using the QIAcuity Software Suite in ~2 hours.
See figures
Applications
The dPCR PanCancer Kits are highly suited for detecting mutations or deletions in DNA isolated from blood, plasma, fresh tissue or FFPE, but other sample types are also possible. The assays are compatible with the QIAGEN workflow and have been validated via wet-lab testing (see figure " Detection of BRAF V600 mutations (A) and EGFR exon 19 deletions (B) in cfDNA extracted from blood plasma using the dPCR PanCancer Kit on the QIAcuity platform"). The results from manual and automated DNA extraction and subsequent dPCR analysis are shown. Data coherence with DNA extracted using different QIAGEN sample preparation methods is demonstrated.
See figures
| Target gene and region | Amino acid position | Nucleotide change | COSMIC ID |
|---|---|---|---|
| dPCR PanCancer Kit BRAF V600 FAM | p.V600K | c.1798_1799delinsAA | COSV56057713 |
| p.V600R | c.1798_1799delinsAG | COSV56058419 | |
| p.V600E | c.1799_1800delinsAA | COSV56059110 | |
| p.V600E | c.1799T>A | COSV56056643 | |
| p.V600D | c.1799_1800delinsAT | COSV56059623 | |
| p.V600G | c.1799T>G | COSV56080151 | |
| p.V600M | c.1798G>A | COSV56075762 | |
| p.V600R | c.1798_1799delinsCG | COSV56288520 | |
| dPCR PanCancer Kit EGFRex19del FAM | p.K745_E749del | c.2233_2247del | COSV51769442 |
| p.E746_A750delinsIP | c.2235_2248delinsAATTC | COSV51817953 | |
| p.E746_A750del | c.2235_2249del | COSV51765119 | |
| p.E746_T751delinsIP | c.2235_2251delinsAATTC | COSV51782151 | |
| p.E746_T751delinsI | c.2235_2252delinsAAT | COSV51850034 | |
| p.E746_A750del | c.2236_2250del | COSV51765066 | |
| p.E746_T751delinsA | c.2237_2251del | COSV51769364 | |
| p.E746_T751delinsV | c.2237_2252delinsT | COSV51775936 | |
| p.E746_T751delinsVA | c.2237_2253delinsTTGCT | COSV51771891 | |
| p.E746_S752delinsV | c.2237_2255delinsT | COSV51765862 | |
| p.L747_A750delinsP | c.2238_2248delinsGC | COSV51782279 | |
| p.L747_T751delinsQ | c.2238_2252delinsGCA | COSV51863059 | |
| p.E746_S752delinsD | c.2238_2255del | COSV51772418 | |
| p.L747_E749del | c.2239_2247del | COSV51780076 | |
| p.L747_A750delinsP | c.2239_2248delinsC | COSV51765099 | |
| p.L747_T751delinsP | c.2239_2251delinsC | COSV51765856 | |
| p.L747_S752del | c.2239_2256del | COSV51767308 | |
| p.L747_S752delinsQ | c.2239_2256delinsCAA | COSV51778874 | |
| p.L747_P753delinsQ | c.2239_2258delinsCA | COSV51785746 | |
| p.L747_T751delinsS | c.2240_2251del | COSV51768180 | |
| p.L747_T751del | c.2240_2254del | COSV51766247 | |
| p.L747_A750delinsS | c.2240_2248del | COSV51810296 | |
| p.L747_P753delinsS | c.2240_2257del | COSV51767961 |
Supporting data and figures
A simple and rapid plate-based workflow
Partitioning, thermocycling and imaging are all integrated into a single fully automated instrument, delivering results in under two hours. Nanoplate formats are amenable to front-end automation for sample preparation, further reducing hands-on time.