✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QIAquick 96 PCR Purification Kit (4)
Cat. No. / ID: 28181
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Up to 95% recovery of ready-to-use DNA
- Fast and convenient procedure
- Cleanup of DNA up to 10 kb in three easy steps
Product Details
QIAquick 96 PCR Purification Kits provide 96-well plates, buffers, and collection tubes for high-throughput silica-membrane-based purification of PCR products >100 bp in size. DNA up to 10 kb is purified using a simple and fast bind–wash–elute procedure and an elution volume of 60–80 µl (resulting in an eluate volume of 40–60 µl). The cleanup procedure can be fully automated on the BioRobot Universal workstation using the QIAquick 96 PCR BioRobot Kit.
Performance
See figures
Principle
QIAquick 96 Kits contain a silica-membrane assembly for binding of DNA in high-salt buffer and elution with low-salt buffer or water. The purification procedure removes primers, nucleotides, enzymes, mineral oil, salts, agarose, ethidium bromide, and other impurities from DNA samples. Silica-membrane technology eliminates the problems and inconvenience associated with loose resins and slurries. Specialized binding buffers are optimized for specific applications and promote selective adsorption of DNA molecules within particular size ranges.
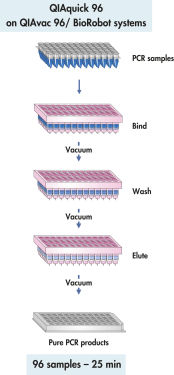
The QIAquick 96 procedure allows parallel purification of up to 96 PCR samples using efficient vacuum-driven purification on a QIAvac 96.
The QIAquick 96 PCR BioRobot Kit is a special kit format optimized for use on the BioRobot Universal. The kit provides QIAquick 96 modules, together with all buffers and plasticware required for automated high-throughput cleanup of 96 PCR samples.
Procedure
The QIAquick system uses a simple bind–wash–elute procedure (see flowchart " QIAquick 96 procedure"). Binding buffer is added directly to the PCR sample or other enzymatic reaction, and the mixture is applied to the 96-well plate. Nucleic acids adsorb to the silica membrane in the high-salt conditions provided by the buffer. Impurities are washed away and pure DNA is eluted with a small volume of low-salt buffer provided or water, ready to use in subsequent applications.
Handling
QIAquick multiwell modules are processed using vacuum-driven purification on QIAvac manifolds. The QIAquick 96 PCR Purification Kit requires the use of the QIAvac 96 vacuum manifold. The cleanup procedure can be fully automated on the BioRobot Uworkstations using the QIAquick 96 PCR BioRobot Kit.
See figures
Applications
DNA fragments purified with the MinElute or QIAquick system are ready for direct use in many applications, including sequencing, microarray analysis, ligation and transformation, restriction digestion, labeling, microinjection, PCR, and in vitro transcription.
Supporting data and figures
QIAquick 96 procedure.
The QIAquick 96 Kit uses a bind-wash-elute procedure with QIAvac 96 or the BioRobot Universal System.
Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 10 µg |
| Processing | Manual/automated |
| Removal <10mers 17–40mers dye terminator proteins | Removal <40mers |
| Sample type: applications | DNA, oligonucleotides: PCR reactions |
| Format | 96-well plate |
| Fragment size | 100 bp – 10 kb |
| Technology | Silica technology |
| Recovery: oligonucleotides dsDNA | Recovery: oligonucleotides, dsDNA |
| Elution volume | 60–80 µl |