✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QuantiTect Virus Kit (1000)
Cat. No. / ID: 211015
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- High sensitivity in single and multiplex assays
- Detection of viral RNA and/or DNA in the same reaction
- Clear detection of weak positive signals
- Fast universal 2-step protocols
- 5x master mix for higher sensitivity with more sample input
Product Details
Performance
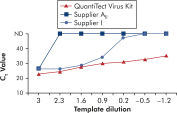
Amplification using QuantiTect Virus Kits provides steep sigmoidal curves over a range of dilutions, even for low template amounts with high CT values (see figure " Unambiguous determination of CT values over a wide dynamic range"). This enables accurate CT value determination for quantification of viral nucleic acids in real-time PCR.
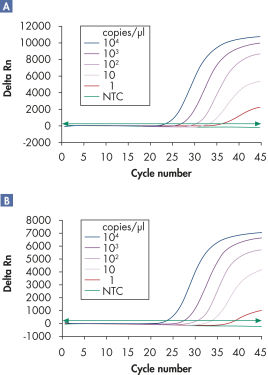
Multiplex assays enable the detection of multiple viral RNA and/or DNA targets plus internal controls over a wide linear range without loss of sensitivity (see figures " Reliable detection of viral RNA over a wide linear range" and " Improved detection of low amounts of viral RNA").
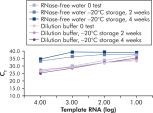
QuantiTect Nucleic Acid Dilution Buffer, supplied with the kits, stabilizes RNA and DNA standards during dilution and reaction setup and prevents loss of nucleic acids on plastic surfaces, such as tubes or pipet tips. The buffer enables reliable dilution of standards used to quantify viral nucleic acids, giving a wide linear range, from low to high CT values and ensures longer storage of standards without degradation (see figure " Reliable dilution and storage of RNA standards").
See figures
Principle
QuantiTect Virus Kits provide highly sensitive detection of viral nucleic acids in single or multiplex assays on the first attempt (see flowchart " QIAGEN multiplex kits"). The optimized master mix ensures that PCR products in a multiplex reaction are amplified with the same efficiency and sensitivity as PCR products in a corresponding single-amplification reaction.
Amplifying control and target genes in the same reaction, instead of separate reactions, increases the reliability of gene quantification by minimizing handling errors. The QuantiTect Virus Buffer contains a balanced combination of K+ and NH4+ ions as well as unique synthetic Factor MP stabilizes, which together promote stable and efficient annealing of primers and probes to the nucleic acid template, enabling high PCR efficiency (see figure " Unique PCR buffer"). In addition, the unique formulation of Sensiscript Reverse Transcriptase ensures highly sensitive reverse transcription of viral RNA, while HotStarTaq Plus DNA Polymerase provides a stringent hot start, preventing the formation of nonspecific products.
| Kit component | Feature | Benefits | |
|---|---|---|---|
| 5x QuantiTect Virus Master Mix | Concentrated master mix | Highly concentrated and optimized for sensitive virus detection | Larger volumes of template can be added to the assay for increased sensitivity |
| HotStarTaq Plus DNA Polymerase | 5 min activation at 95ºC | Set up of qPCR reactions at room temperature | |
| QuantiTect Virus Buffer | Balanced combination of NH4+ and K+ ions | Specific primer annealing ensures reliable PCR results | |
| Synthetic Factor MP | Reliable multiplexing analysis of up to 4 genes in the same tube | ||
| Additional kit components | QuantiTect Virus RT Mix | Contains a unique formulation of Sensiscript Reverse Transcriptase | Optimized for highly sensitive detection of viral RNA |
| QuantiTect Nucleic Acid Dilution Buffer | Proprietary buffer formulation for dilution and storage of nucleic acid standards. | Stabilizes RNA and DNA standards during dilution and reaction setup and prevents loss of nucleic acids on plastic surfaces, such as tubes or pipet tips |
See figures
Procedure
QuantiTect Virus Kits provide highly sensitive real-time PCR analysis of viral nucleic acids (RNA and/or DNA) and internal controls using sequence-specific probes. Reactions can be carried out with or without a reverse-transcription step, enabling flexible design of multiplex assays to detect RNA targets, DNA targets, or both RNA and DNA targets. Follow the protocol in the handbook for fast and reliable results.
Kits are available with or without ROX passive reference dye in the master mix (see table).
| ROX dye | Kit | Compatible cyclers |
|---|---|---|
| Supplied in master mix | QuantiTect Virus Kit | All cyclers from Applied Biosystems except Applied Biosystems 7500 |
| Supplied in separate tube | QuantiTect Virus +ROX Vial Kit | Applied Biosystems 7500 and cyclers from Bio-Rad, Cepheid, Eppendorf, QIAGEN, Roche, Agilent, and other suppliers |
For fast and highly sensitive end-point one-step RT-PCR applications, including virus detection, we recommend using the QIAGEN OneStep RT-PCR Kit.
Applications
Supporting data and figures
Unambiguous determination of CT values over a wide dynamic range.
Specifications
| Features | Specifications |
|---|---|
| Applications | Virus detection |
| SYBR Green I or sequence-specific probes | Sequence-specific probes |
| Real-time or endpoint | Real-Time |
| Reaction type | Reverse Transcription and PCR |
| Thermal cycler | Most real-time cyclers (except capillary cyclers e.g. LightCycler® 1.x and 2.0) |
| Sample/target type | RNA and/or DNA targets |
| With or without ROX | Available with ROX in master mix and ROX as a separate vial |
| Single or multiplex | Single or multiplex |