miRCURY LNA miRNA Probe PCR Assay (200)
Cat. No. / ID: 339350
Features
- LNA-enhanced, miRNA-specific forward PCR primer and hydrolysis probe
- Detect true signals at very low detection range, down to 1 miRNA copy
- Fast and simple two-step workflow that takes 2 hours
- High level of robustness and reproducibility for reliable results
Product Details
miRCURY LNA miRNA Probe PCR Assays are miRNA PCR primer-probe sets that enable extremely sensitive and specific miRNA quantification with the miRCURY LNA miRNA Probe PCR System. Robust and sensitive, the system enables accurate detection of miRNA from samples with low RNA content, including serum, plasma and urine. Both the forward PCR amplification primer and the FAM- and dark quencher-labeled probe are miRNA-specific and optimized with LNA technology, which boosts assay performance. Assays for all-important miRNAs are wet-bench validated. Next to the predesigned assays, which cover all miRNAs in miRBase, miRCURY LNA miRNA Custom Probe PCR Assays can be created easily for any miRNA of interest using the same design algorithm.
Need a quote for your research project or would you like to discuss your project with our specialist team? Just contact us!
Performance
Highly sensitive detection of true signals
The exceptional sensitivity of miRCURY LNA miRNA Probe Assays is achieved by combining universal reverse transcription with LNA-enhanced, Tm-normalized, miRNA-specific primers and probe. This combination enables accurate and reliable quantification of individual miRNAs from as little as 1 pg total RNA or down to 1 molecule without pre-amplification (see Accurate quantitation even with low starting material and exceptional efficiency across a broad dynamic range). The sensitivity and quick workflow make the system particularly well-suited for serum or plasma samples and the QuantiNova enzymes ensure highly reproducible results between technical and biological replicates at the first attempt, saving sample material and time.
Compared with other miRNA probe-based PCR systems that rely on only the probe to generate specificity, the LNA-enhanced primer and probe of the miRCURY LNA miRNA Probe Assays are both miRNA-specific and short enough to prevent overlap when binding to the target (see miRCURY LNA miRNA Probe PCR System at a glance). As a result, miRCURY LNA miRNA Probe Assays outperform alternative solutions in terms of efficiency (see Improved efficiency compared to alternative solutions), linearity and sensitivity without pre-amplification (see Improved linearity and high sensitivity without pre-amplification). Combined with excellent signal-to-noise ratio (see Accurate detection even with high RNA background), this unique feature allows discriminating closely related miRNAs with sequence differences as little as one nucleotide (see Discriminatory power down to a single nucleotide difference).
Validated and optimized
Assay designs (primer and probe) are based on a design algorithm created by LNA and assay experts. This comprehensive algorithm evaluates and optimizes 60 parameters that are validated in silico against highly stringent performance criteria to guarantee a design that offers the highest sensitivity. The outcomes are assays able to detect down to 1 miRNA copy. Assays for all important miRNAs are also fully wet-bench validated.
Coverage
Over 30,000 assays are available covering all miRBase 22. Over 1,000 assays are fully wet-lab validated for testing sensitivity, specificity, efficiency and background. The remaining assays are in silico-validated using a comprehensive design algorithm with stringent assay performance criteria that ensures high-quality, species-specific, LNA-enhanced assays with optimal sensitivity and specificity for each organism. If you are working with novel miRNAs, such as from an NGS experiment, design your own assay with the miRCURY LNA miRNA Custom Probe PCR Assays.
Fast and easy
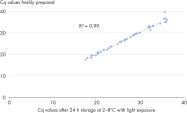
The easy-to-follow, 2-hour protocol saves time and labor. By using a universal transcription as well as optimized RT-PCR protocols, the procedure is greatly simplified compared to other solutions. Furthermore, the cDNA generated in the RT step can be used across the miRCURY LNA miRNA Probe PCR System, allowing you to seamlessly transition from panels to assays depending on your research needs, and saving sample material and time. The number of pipetting steps is reduced to a minimum, which also minimizes technical variation. Assay robustness allows room temperature setup and delayed runs for greater flexibility and use in automation (see Reaction stability and setup robustness). This simplified workflow coupled to ISO-compliant assay manufacturing (see High lot-to-lot consistency) makes it possible to achieve extremely high day-to-day and site-to-site reproducibility.
See figures
 miRCURY LNA miRNA Probe PCR System at a glance.
miRCURY LNA miRNA Probe PCR System at a glance. Improved efficiency compared to alternative solutions.
Improved efficiency compared to alternative solutions. Improved linearity and high sensitivity without pre‐amplification.
Improved linearity and high sensitivity without pre‐amplification. Accurate detection even with high RNA background.
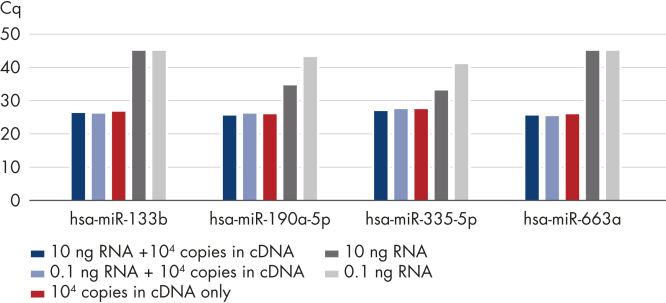
Accurate detection even with high RNA background. Discriminatory power down to a single nucleotide difference.
Discriminatory power down to a single nucleotide difference. High lot-to-lot consistency.
High lot-to-lot consistency.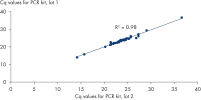
Principle
The miRCURY LNA miRNA Probe PCR System enables sensitive, specific miRNA quantification and profiling via real-time PCR. Detection is based on a FAM- and dark quencher-labeled probe quencher-labeled hydrolysis probe. The system covers all steps involved in converting miRNA to cDNA and subsequent qPCR detection of miRNAs. The miRCURY LNA miRNA Probe PCR Assays have been validated using the QuantiNova Probe Master Mix; use of other reagents for qPCR will affect the results.
RNA Spike-In Kit for RT
The RNA Spike-In Kit enables quality control of the RNA isolation, cDNA synthesis and PCR amplification steps of miRCURY LNA miRNA Probe PCR experiments so you can identify any poor-quality samples early in the process and save time and reagents. The kit includes UniSp2, UniSp4 and UniSp5 RNA Spike-in Template Mix and the cel-miR-39-3p RNA Spike-in Template.
miRCURY LNA RT Kit
One universal reverse transcription kit for two detection systems. The miRCURY LNA RT Kit is used for both the miRCURY LNA miRNA PCR System that employs SYBR Green for detection, and the probe-based miRCURY LNA miRNA Probe PCR System. The kit includes two system-specific reaction buffers and all the necessary reagents to perform fast and convenient miRNA polyadenylation and reverse transcription in a single reaction step.
miRCURY LNA Probe PCR Kit
Optimized and dedicated for the miRCURY LNA miRNA Probe PCR System, this kit contains the high-performance QuantiNova Probe Master Mix and the miRCURY Probe Universal Reverse Primer. Combined with a miRCURY LNA miRNA Probe PCR Assay or a miRCURY LNA miRNA Probe PCR Panel, the chemistry enables miRNA detection via quantitative real-time PCR.
miRCURY LNA miRNA Probe PCR Assays
These primer and probe assays use miRNA-specific hydrolysis probes and miRNA-specific forward primers designed to detect mature miRNAs using the most up-to-date sequence information from miRBase 22. The miRCURY Probe Universal Reverse Primer is contained in the miRCURY Probe PCR Kit.
miRCURY LNA miRNA Probe PCR Panels
Ready-to-use, predesigned, single-use PCR panel plates to rapidly profile mature miRNAs focusing on the miRNome, specific pathways, certain sample types like serum/plasma, or in a user-designed custom panel.
GeneGlobe Data Analysis Center
This complimentary, web-based analysis tool guides you step-by-step via an easy wizard through the normalization and analysis of your data and generates publication-ready figures.
Spike-in assays
| Product name | Control type | Used to control | Targeted template |
| UniSp2 miRCURY LNA miRNA Probe Assay | Spike-in control assay | Extraction efficiency | UniSp2 – synthetic RNA template, incl. in RNA Spike-In Kit, for RT – 339390 |
| UniSp4 miRCURY LNA miRNA Probe Assay | Spike-in control assay | Extraction efficiency | UniSp4 – synthetic RNA template, incl. in RNA Spike-In Kit, for RT – 339390 |
| UniSp5 miRCURY LNA miRNA Probe Assay | Spike-in control assay | Extraction efficiency | UniSp5 – synthetic RNA template, incl. in RNA Spike-In Kit, for RT – 339390 |
| UniSp6 miRCURY LNA miRNA Probe Assay | RT control assay | Reverse transcription reaction | UniSp6 – synthetic RNA template, included in miRCURY LNA RT Kit – 339340 |
| UniSp9 miRCURY LNA miRNA Probe Assay | IPC – interplate calibrator | PCR reaction | UniSp9 – synthetic DNA template. Spotted with assay on the plate. Only available on plates (panels) |
| cel-miR-39-3p miRCURY LNA miRNA Probe Assay | RT control assay | Reverse transcription reaction | UniSp6 – synthetic RNA template, incl. in RNA Spike-In Kit, for RT – 339390 |
Control assays
| Product name | Target organism | Alternative names | NCBI reference |
| Control primer set, SNORD38B (hsa) | hsa | U38B; RNU38B | NR_001457 |
| Control primer set, SNORD44 (hsa) | hsa | U44; RNU44 | NR_002750 |
| Control primer set, SNORD48 (hsa) | hsa | U48; RNU48 | NR_002745 |
| Control primer set, SNORD49A (hsa) | hsa | U49; U49A; RNU49 | NR_002744 |
| Control primer set, SNORA66 (hsa) | hsa | U66; RNU66 | NR_002444 |
| Control primer set, 5S rRNA (hsa) | hsa, mmu | V00589 | |
| Control primer set, U6 snRNA (hsa, mmu) | hsa, mmu, rno | U6 | X59362 |
| Control primer set, RNU5G snRNA (mmu, hsa) | mmu, hsa, rno | Rnu5a; U5a; Rnu5g | NR_002852 |
| Control primer set, RNU1A1 (mmu, hsa) | mmu, hsa, rno | Rnu1a-1; Rnu1a1 | NR_004411 |
| Control primer set, SNORD65 (mmu) | mmu | U65; RNU65 | NR_028541 |
| Control primer set, SNORD68 (mmu) | mmu | U68; RNU68 | NR_028128 |
| Control primer set, SNORD110 (mmu) | mmu | U110; RNU110 | NR_028547 |
Procedure
The first step, the reverse transcription reaction, is an easy-to-perform, single-tube reaction. Total RNA, including miRNA, is used as starting material and the resulting cDNA is used in the real-time PCR amplification with LNA-enhanced primers and hydrolysis probe to detect the mature miRNAs. This enables straightforward miRNA quantification and profiling, accessible to any lab with a real-time PCR cycler.
Applications
The miRCURY LNA miRNA Probe PCR Assays are used as part of the miRCURY LNA miRNA Probe PCR System for:
- miRNA expression detection, profiling and quantification
- miRNA biomarker development – screening and identification
- miRNA regulation and pathway analysis
- miRNA detection in complex sample types and samples with low miRNA expression levels (e.g., exosomes, serum, plasma, urine, FFPE sections)
- snoRNA detection
- Validation of miRNA sequencing
Supporting data and figures
Accurate detection even with high RNA background.