✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
REPLI-g FFPE Kit (25)
Cat. No. / ID: 150243
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Whole genome amplification directly from paraffin sections
- Whole genome amplification of purified DNA from FFPE tissue
- Scalable and standardized DNA yields: up to 40 µg per tissue section
- Fast and easy protocol for rapid results
Product Details
A lack of sufficient quantities of genomic DNA for genomic analysis can be overcome by global amplification of all DNA within a sample (whole genome amplification). The REPLI-g FFPE Kit consists of DNA Polymerase, novel buffers, and reagents that enable efficient whole genome amplification from formalin-fixed and paraffin embedded (FFPE) tissue without the requirement for prior DNA purification. After lysis of the tissue section, the DNA is processed so that fragmented DNA is ligated. The long DNA strands created are amplified using proven REPLI-g technology.
Performance
For the two protocols with different amplification times, typical yields per 50 µl reaction with standard quality templates are up to 10 µg after 2-hour amplification (standard reaction) and up to 40 µg after 8-hour amplification (high-yield reaction).
Principle
The availability of sufficient quantities of genomic DNA for genomic analysis is often lacking. Whole genome amplification (WGA) overcomes this limitation by global amplification of all DNA within a sample, providing sufficient quantities to perform all analyses on the same DNA sample.
Genotyping of formalin-fixed, paraffin-embedded (FFPE) tissue samples enables morphological tissue changes to be directly linked to specific genome mutations. However, formalin fixation causes irreversible damage to the DNA, resulting in fragmented DNA that is cross-linked to other biomolecules within the sample. In addition, the limited amount of DNA that can be extracted from FFPE tissue samples is only sufficient for a few analyses.
The REPLI-g FFPE Kit overcomes these problems through a procedure involving a novel DNA processing reaction that prepares and ligates fragmented DNA (see figure " REPLI-g FFPE principle"). Whole genome amplification of this randomly ligated DNA is then performed using proven REPLI-g technology. REPLI-g products combine multiple displacement amplification (MDA) with a uniquely processive DNA polymerase. Due to the high processivity and strand displacement activity, REPLI-g DNA Polymerase minimizes unequal sequence and locus representation. This provides much more reliable results when compared to PCR-based WGA methods. The novel DNA processing reaction allows the advantages of MDA to be extended to highly degraded DNA samples derived from FFPE tissue. Precious sample material can be amplified whilst maintaining locus representation, enabling unlimited downstream analyses to be performed.
See figures
Procedure
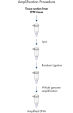
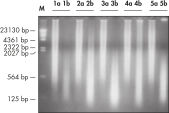
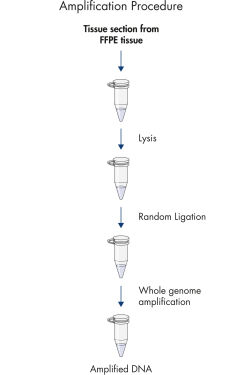
The REPLI-g FFPE protocol allows the amplification of purified DNA from FFPE tissue samples or DNA direct from FFPE tissue samples without prior DNA purification. After lysis of the tissue section, the DNA is processed using novel buffers and enzymes that ligate fragmented DNA (see flowchart " REPLI-g FFPE procedure"and figure " High-molecular-weight ligated DNA"). The long DNA strands created by the ligation reaction are amplified using proven REPLI-g technology. Once amplified, the DNA is suitable for immediate use in most downstream genotyping assays without further purification.
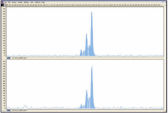
DNA amplified using the REPLI-g FFPE procedure is highly suited for use in real-time PCR (e.g., using QuantiFast Kits) and end-point PCR (e.g., using the QIAGEN Fast Cycling PCR Kit), with a PCR amplicon preferably smaller than the average fragment size of the starting template. Further applications are microsatellite analysis (see figure " Reliable microsatellite analysis") and SNP genotyping (REPLI-g FFPE amplified DNA may not be suited for genotyping methods that require restriction digestion for DNA labeling).
See figures
Applications
DNA amplified using the REPLI-g FFPE procedure is highly suited for use in:
- Real-time PCR
- End-point PCR
- SNP genotyping
- Microsatellite analysis
Supporting data and figures
REPLI-g FFPE procedure.
Specifications
| Features | Specifications |
|---|---|
| Yield | 10 µg (2h reaction)-40 µg (8h reaction) |
| Quality assessment | No |
| Starting material | FFPE tissue sections or purified genomic DNA |
| Technology | MDA |
| Maximum input volume | 1 tissue section (10-40 µm thickness) |
| Reaction time | 2-8h (2h: standard; 8h: high-yield reaction) |
| Reaction volume | 50 µl |
| Starting amount of DNA | 100-300 ng* |
| Applications | Genotyping, PCR, Real-time PCR |
| Samples per run (throughput) | medium |
| Amplification | Whole genomic DNA |