✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QIAseq Targeted RNA Panel Human TCR (12)
Cat. No. / ID: 334651
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Unique molecular indexing (UMI) technology removes amplification, duplication and sequencing artifacts to increase accuracy
- Profile single TCR receptors (TCR-alpha, TCR-beta, TCR-gamma, TCR-delta) or combine panels for a comprehensive immune repertoire
- Single-tube library construction and versatile panels allow analysis of one, two, three or all four receptors at once
- Unique dual indexing (UDI) minimizes sample index misassignment
- Includes access to GeneGlobe Analyze for data analysis, including read alignment and clonotype report
Product Details
The QIAseq Targeted RNAseq Panel for T-cell Receptor enables targeted next-generation sequencing (NGS) of the human or mouse expressed T-cell receptor (TCR), which includes four distinct genes: TCR-alpha, TCR-beta, TCR-gamma and TCR-delta. This highly optimized solution incorporates unique molecular indices (UMIs) to facilitate ultrasensitive and accurate characterization of the immune repertoire in cells and tissues starting from 200 pg to 1000 ng of total RNA.
The adaptive immune system is composed of T and B lymphocytes that bind antigens via highly specific T-cell receptors (TCRs) and B cell receptors (BCRs) on their cell surfaces. To recognize a nearly infinite number of potential antigens, extensive sequence diversity of TCRs and BCRs is generated by somatic V(D)J recombination of the TCR and BCR loci, and by subsequent somatic hypermutation and class switch recombination of the BCR upon antigen stimulation. Accurate characterization of the TCR and BCR repertoires is key to understanding adaptive immune responses and has many applications across different fields, including vaccine development, autoimmunity, monitoring treatment response in lymphoid malignancies and immunotherapy.
Compared to traditional methods, NGS provides an unprecedented, high-resolution picture of the immune repertoire. However, many available immune repertoire sequencing methods involve multiplex PCR with primers targeting different V or J regions, which can introduce substantial amplification bias.
The QIAseq Targeted RNAseq Panel for T-cell Receptor utilizes unique molecular indices (UMIs) with QIAseq Enrichment Technology to robustly create targeted RNA-seq libraries for NGS instruments. This library construction approach greatly improves amplification uniformity compared to multiplexed PCR-based V and J primer pools. The incorporation of UMIs before the amplification step further reduces amplification bias and allows for accurate and sensitive TCR clonotype and repertoire diversity assessment.
The QIAseq Targeted RNAseq Panel for T-cell Receptor is a complete Sample to Insight solution for precise characterization of the TCR immune repertoire using NGS. The purchase of the TCR Panel includes access to GeneGlobe Analyze, which contains a highly optimized pipeline for read mapping and clonotype calling.
Principle
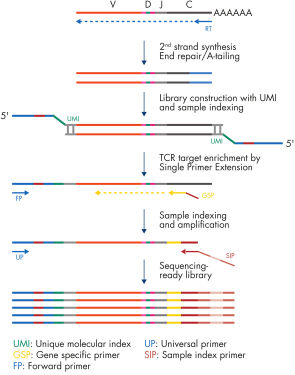
The QIAseq Targeted RNAseq Panel for T-cell Receptor provides accurate and sensitive TCR clonotype and diversity assessment using QIAGEN Enrichment Technology. TCR-specific primers for reverse transcription and enrichment are provided along with all the necessary library construction reagents. The QIAseq Targeted RNAseq Panel for T-cell Receptor is designed to enrich TCR α, β, γ and σ subunits (individually or combined) in a single reaction using 200 pg to 1000 ng of RNA (see figure "QIAseq Targeted RNAseq Panel for T-cell Receptor workflow").
See figures
Procedure
cDNA synthesis
Start with total RNA from peripheral blood leukocytes, whole blood, enriched T-cells or solid tumors. RNA samples are first reverse-transcribed into cDNA with gene-specific primers. Subsequently, second-strand synthesis occurs, which generates double-stranded cDNA (ds-cDNA). This ds-cDNA is then end-repaired with an A-addition in a single-tube protocol.
Unique Molecular Index (UMI) assignment
Prior to target enrichment and library amplification, each original cDNA molecule is assigned a unique molecular index (UMI) containing up to 18 bases. This is achieved by ligating a random sequence adapter containing the UMI to the ds-cDNA. Statistically, this process provides >268 million possible indices per adapter, with each DNA molecule in the sample receiving a unique UMI sequence.
Target enrichment and final library construction
Following UMI assignment, QIAseq Enrichment Technology is performed to capture TCR cDNA molecules with UMIs into the NGS library. For enrichment, ligated cDNA molecules are subjected to a TCR-specific enrichment using a TCR targeted panel. After enrichment, a universal PCR is performed to amplify the library and introduce unique dual indices.
NGS adapter and Unique Dual Index (UDI) technologies
The QIAseq Targeted RNAseq Panel for T-cell Receptor requires the QIAseq 24-Index TCR UDI (24) (cat. no. 334792) or QIAseq 96-Index TCR UDI Set A (96) (cat. no. 334805), which contain the TUDI adapters and dual index primers. The UDI design significantly reduces the risk of index bleeding issues (“index hopping”) associated with next-generation sequencing instruments that utilize patterned flow cells. With unique dual indexing (UDI), each sample will be assigned two unique sample indices to overcome the error introduced by image analysis, sequencing error and demultiplexing, and to remove misassignment of sequencing data to the wrong samples.
Next-generation sequencing on Illumina NGS systems
The QIAseq Targeted RNAseq Panel for T-cell Receptor is compatible with Illumina NGS systems (MiSeq, NextSeq 1000/2000 and NovaSeq 6000). The best sequencing results are achieved using 500 or 600 cycle flow cells.
Data analysis
The QIAseq Immune Repertoire pipeline automatically performs all steps necessary to generate TCR diversity and clonotype reports from your raw NGS data. The QIAseq Immune Repertoire pipeline is available through cloud, desktop and server software packages. For cloud-based analysis, researchers can utilize the QIAseq section of GeneGlobe Analyze. For desktop and server support, QIAGEN Genomics Workbench can be used.
Sample specifications |
|
|---|---|
| RNA types | Total RNA, FFPE/fragmented RNA and enriched mRNA |
| Type of library | Targeted RNA-seq with UMIs |
| Gene targets | Human: TRA, TRB, TRD, TRG Mouse: Tcra, Tcrb, Tcrd, Tcrg |
| Minimum concentration of RNA | 40 pg/µL |
| Volume of RNA per reverse transcription reaction | 5 µL |
| Total volume of cDNA reaction | 6 µL |
| RIN requirements | 1 to 10 |
| Minimum DV 600% | 30% |
Sequencing specifications |
Research goal |
|
|---|---|---|
| Sample type | Survey with shallow sequencing | Comprehensive with deep sequencing |
| Peripheral blood leukocytes: 200 pg to 1000 ng of total RNA | 0.016–80 M reads | 0.32–1600 M reads |
| Whole blood: 500 pg to 200 ng of total RNA | 0.01–4 M reads | 0.2–80 M reads |
| Tumor tissue: 100 ng to 1000 ng of total RNA | 0.12–1.2 M reads | 1.2–12 M reads |
| Purified T-cells: 10 to 10,000 cells of RNA | 0.002–2 M reads | 0.04–40 M reads |
Product specifications |
|
|---|---|
| Supported index kits (required) | 334792 QIAseq 24-Index TCR UDI (24) 334805 QIAseq 96-Index TCR UDI Set A (96) |
| Recommended control RNA for monitoring library performance (sold separately) | QIAGEN XpressRef Universal Total RNA |
| Recommended library quantification kits (sold separately) | QIAseq Library Quant Assay Kit Cat. No. / ID: 333314 |
| Online data analysis included with kit | GeneGlobe Analyze |
| Standalone desktop data analysis (sold separately) | CLC Genomics Workbench |
| Recommended NGS instruments | Illumina MiSeq or NextSeq 1000/2000 |
| Recommended sequencing cycles/length | Paired-end, 600 cycles on Illumina NextSeq 1000/2000 |
Supporting data and figures
QIAseq Targeted RNAseq Panel for T-cell Receptor workflow