QIAsure Methylation Test Kit
For triage testing of women who are high-risk HPV positive or have ASC-US cytology
For triage testing of women who are high-risk HPV positive or have ASC-US cytology
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 616014
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
The QIAsure Methylation Test is a multiplex real-time methylation-specific PCR (qMSP)-based assay for the detection of promoter hypermethylation of the genes FAM19A4 and hsa-mir124-2. Samples that may be tested with QIAsure Methylation Test include bisulfite-converted DNA isolated from physician-collected cervical specimens or self-collected vaginal samples. Want to try this solution for the first time? Request a quote for a trial kit.
Self-screen B.V. is the legal manufacturer of the QIAsure Methylation Test.
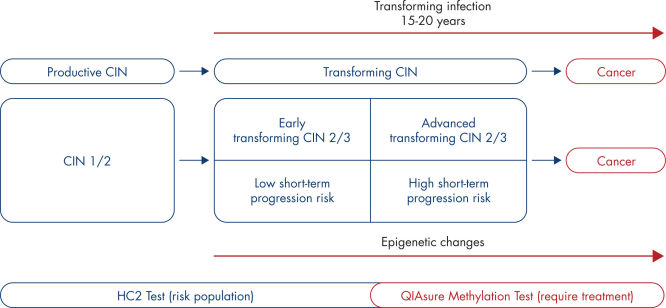
The presence of high-risk HPV in epithelial cells in the cervix may result in transforming lesions in some women and hence development of cervical cancer (1–9). However, HPV is a common infection and in most cases it does not cause a pre-cancerous or cancerous lesion and is simply cleared by the woman’s immune system. Increased expression of the viral oncogenes E6 and E7 in proliferating epithelial cells drives HPV induced carcinogenesis and results in a transforming HPV infection. The associated cervical precursor lesion, also called transforming cervical intraepithelial lesion or transforming CIN, may ultimately progress to cervical cancer. (See Natural history of the development of cervical cancer).
This process is associated with the accumulation of epigenetic alterations, i.e., increased DNA methylation in specific tumor suppressor genes. By detecting promoter hypermethylation of the tumor suppressor genes FAM19A4 and/or hsa-mir124-2, women with advanced transforming CIN and hence at a high risk of short-term disease progression can be distinguished from women with productive or early transforming CIN who are at low risk of progression to cancer. (See Heterogeneity in CIN and associated risk of disease progression).
The QIAsure Methylation Test is a multiplex real-time PCR test that amplifies the methylated promoter regions of the tumor suppressor genes FAM19A4 and hsa-mir124-2, as well as a methylation-unspecific fragment of a reference gene that acts as an internal sample quality control. The QIAsure assay runs on Rotor-Gene Q MDx instrument and the Rotor-Gene AssayManager software automatically performs data analysis and interpretation. (See QIAsure Methylation Test).
The QIAsure Methylation Test can be used to identify hypermethylation of genes FAM19A4 and hsa-mir124-2 and is a follow-up test for women with a positive HPV DNA test. It is also indicated for use in women who have a Pap smear showing atypical squamous cells of undetermined significance (ASC-US). QIAsure Methylation Test is able to detect CIN 3 at a high risk of short-term progression and cancerous cells with a higher sensitivity compared with cytology or HPV 16/18 genotyping (8). As the QIAsure Methylation Test also has low sensitivity for CIN with low short-term progression risk, it can be used in triage to distinguish women who would benefit from increased surveillance from those who need immediate colposcopy.>
| Clinical endpoint | Fraction | Positivity rate, % (95% CI) |
|---|---|---|
| ≤CIN 1 | 24/117 | 20.5 (14.1–28.8) |
| CIN 2 | 16/42 | 38.1 (24.8–53.4) |
| CIN 3 | 20/30 | 66.7 (48.4–84.0) |
| Squamous cell carcinoma | 59/59 | 100.0 (94.0–100.0) |
| Adenocarcinoma | 10/10 | 100.0 (69.0–100.0) |
| CIN 3+* | 89/99 | 89.9 (82.2–94.5) |
| All cervical carcinoma*† | 69/69 | 100.0 (94.0–100.0) |
| Clinical endpoint | Fraction | Positivity rate, % (95% CI) | ||
|---|---|---|---|---|
| ≤CIN 1 | 34/148 | 23.0 (16.9–30.4) | ||
| CIN 2 | 7/24 | 29.2 (14.6–49.8) | ||
| CIN 3 | 33/50 | 66.0 (52.0– 77.7) | ||
| Squamous cell carcinoma | 8/8 | 100.0 (63.1–100.0) | ||
| Adenocarcinoma | 3/3 | 100.0 (29.2–100.0) | ||
| CIN 3+* | 44/61 | 72.1 (59.7–81.9) | ||
| All cervical carcinoma*† | 11/11 | 100.0 (72.0–100.0) | ||
References
1. De Strooper, L.M., et al. (2014) Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev. Res. 7, 1251–7.
2. Bierkens, M. et al. (2013) CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease Int. J. Cancer 133, 1293–9.
3. Costello, J.F., and Plass, C. (2001) Methylation matters. J. Med. Genet. 38, 285–303.
4. Wilting, S.M., et al. (2010) Methylation-mediated silencing and tumour suppressive function of has-MiR-124 in cervical cancer. Mol. Cancer 9, 167.
5. De Strooper, L.M., et al. (2014) CADM1, MAL and miR12-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J. Clin. Pathol. 67, 1067–71.
6. De Strooper, L.M., et al. (2016) Comparing the performance of FAM19A4 methylation analysis, cytology and HPV 16/18 genotyping for the detection of cervical (pre)cancer in high-risk HPV-positive women of a gynecologic outpatient population (COMETH study). Int. J. Cancer 138, 992–1002.
7. De Strooper, L.M., et al. (2016) Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre)cancer in HPV-positive women. Gynecol. Oncol. 141, 341–7.
8. Steenbergen R. D.M., et al (2014). Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat. Rev. Cancer 14, 395–405.
9. Luttmer R., et al. (2016). Management of high-risk HPV-positive women for detection of cervical (pre)cancer. Expert Rev. Mol. Diagn. 16(9), 961–74.